Proteomics in multiple sclerosis
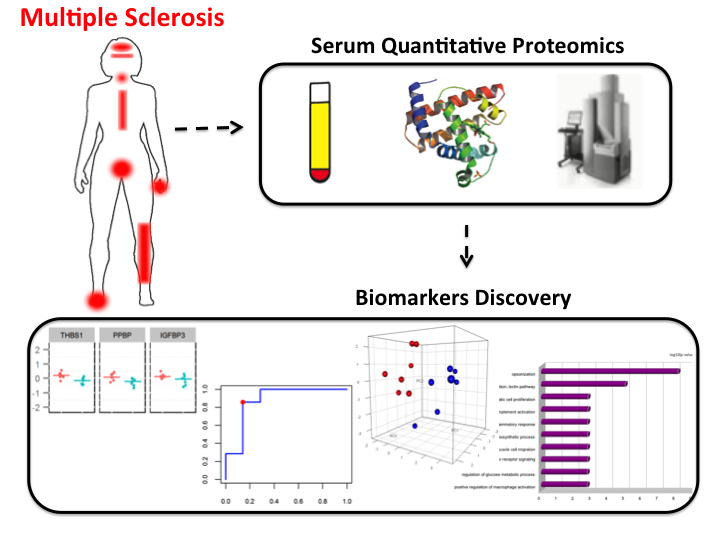 I have collaborated with Dr. Helen Tremlett, from the Pharmacoepidemiology in Multiple Sclerosis Research Group (UBC), working on a proteomics study of Multiple Sclerosis. Using advanced technological methods and an elastic net model, we identified and built a score based on a panel of 11 proteins to classify samples from MS patients into two extreme MS phenotypic groups (benign and aggressive cases of MS) with high estimated sensitivity and specificity. Our study was one of the first ones to explore the joint association of multiple proteins with MS disease progression serum samples. Image from Tremlett et al., Journal of Proteomics 118, 2-11. PubMed
I have collaborated with Dr. Helen Tremlett, from the Pharmacoepidemiology in Multiple Sclerosis Research Group (UBC), working on a proteomics study of Multiple Sclerosis. Using advanced technological methods and an elastic net model, we identified and built a score based on a panel of 11 proteins to classify samples from MS patients into two extreme MS phenotypic groups (benign and aggressive cases of MS) with high estimated sensitivity and specificity. Our study was one of the first ones to explore the joint association of multiple proteins with MS disease progression serum samples. Image from Tremlett et al., Journal of Proteomics 118, 2-11. PubMed
Genetic mutations in DLBCL
My research group is currently collaborating with Dr. Randy Gascoyne, Dr. David Scott, and Dr. Sohrab Shrab from the BC Cancer Agency on a genetic study of diffuse large B-cell lymphoma (DLBCL). In this work, we identify genetic alterations driving two distinct cell-of-origin (COO) subtype-specific tumor growth, providing a deeper understanding of DLBCL pathogenesis. In particular, we proposed an extension of the Cox-LASSO model to build a prognostic genetic signature of time to cancer progression beyond the signal that correlates with COO. The resulting genetic score improves the prediction of survival compared to the prediction based on COO alone. A manuscript summarizing the results of this collaboration is under review at Nature Genetics.
Proteomics and genomics in allergic asthma
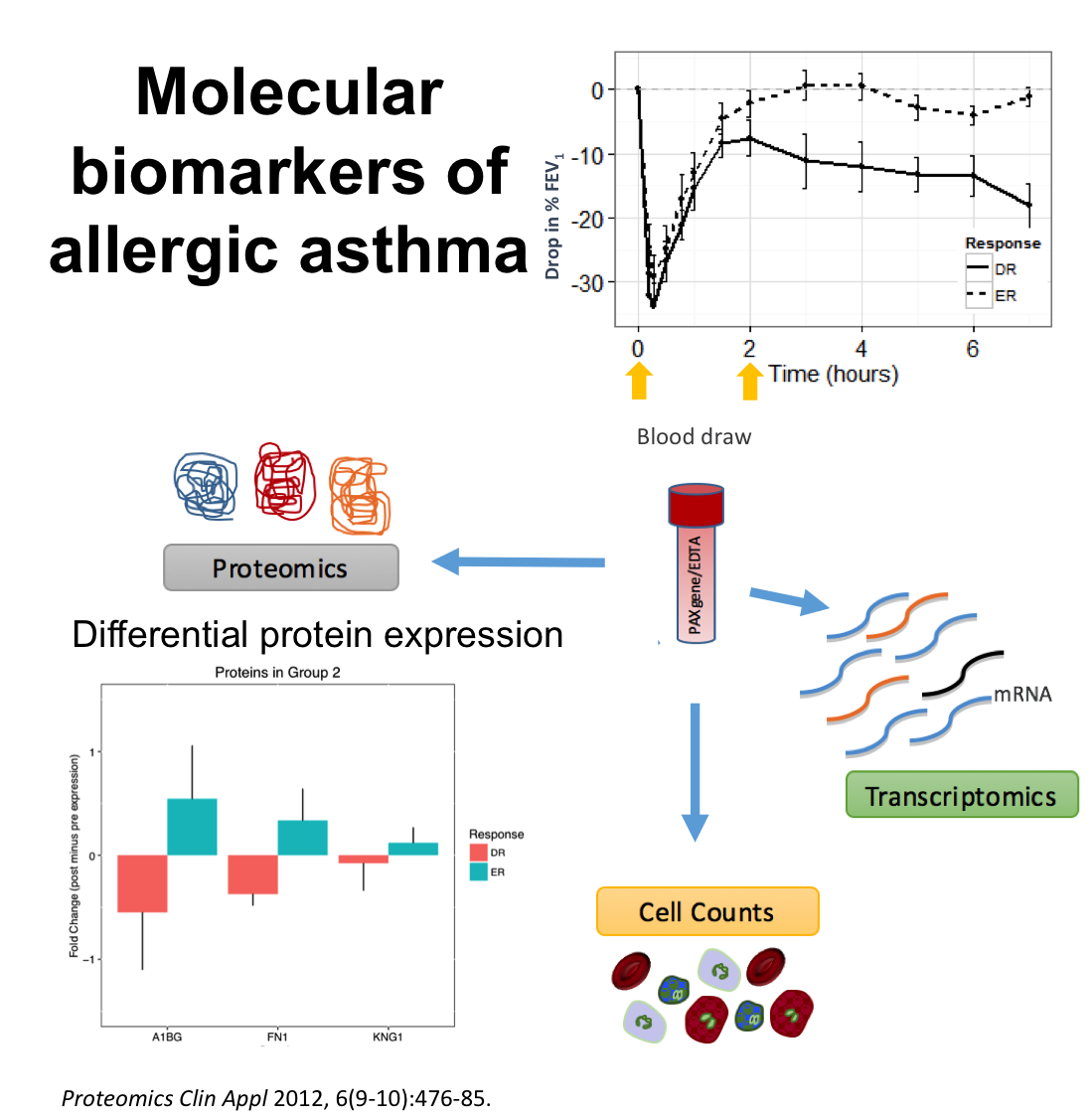 Together with Dr. Scott Tebbutt, from iCAPTURE and PROOF Centre, we are working on the analysis of genomics and proteomics data from patients with allergic asthma. Through this collaboration we have built a comprehensive analytical pipeline to identify and validate molecular markers of different asthmatic phenotypes in order to characterize the molecular mechanisms related to allergic asthma. Image (partial) from Singh et al. (2012) Proteomics Clinical Applications 6, 476-485.
PubMed
Together with Dr. Scott Tebbutt, from iCAPTURE and PROOF Centre, we are working on the analysis of genomics and proteomics data from patients with allergic asthma. Through this collaboration we have built a comprehensive analytical pipeline to identify and validate molecular markers of different asthmatic phenotypes in order to characterize the molecular mechanisms related to allergic asthma. Image (partial) from Singh et al. (2012) Proteomics Clinical Applications 6, 476-485.
PubMed
Proteomics in cardiovascular diseases
I have worked with Dr. Bruce McManus, from the NCE CECR Prevention of Organ Failure (PROOF) Centre of Excellence (UBC), and Dr. Christoph Borchers, from the Genome BC Proteomics Centre (University of Victoria), on proteomic biomarkers studies of heart failure and cardiovascular diseases (work in progress). We have conducted rigorous statistical analyses based on regularized linear models and classifiers that identified promising panels of proteins. If externally validated, these markers will lead to the development of inexpensive protein-based blood tests that could be used by physicians to monitor response to therapies, resulting in more personalized patient management.
Proteomics in transplantation
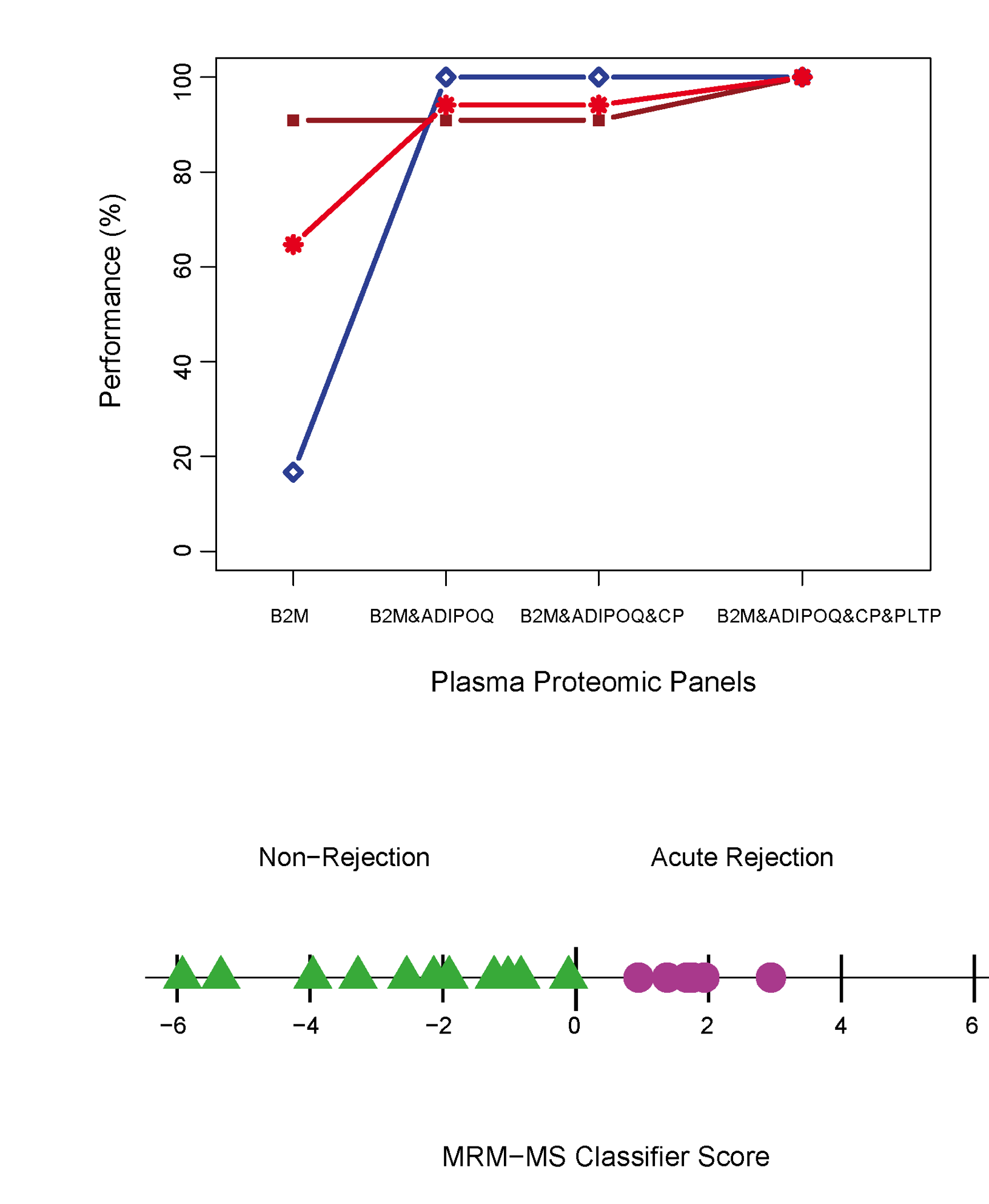 Since 2005, I have been working in the realm of molecular biomarker discoveries in Transplantation through my participation in the Genome Canada-funded Biomarkers in Transplantation initiative and with the NCE CECR Prevention of Organ Failure (PROOF) Centre of Excellence. In more recent collaborations with the group, I have developed a computational proteomic pipeline founded on the combination of advanced statistical methodologies to identify, validate, and clinically implement biomarker blood-based solutions to diagnose and predict various diseases. We demonstrated the utility of the proposed methodologies in various biomarkers studies of cardiac transplantation. The computational methodologies proposed in this pipeline provide a reference to a wide range of biomarker proteomic studies. Image from Cohen Freue et al. PLoS Comp Biol 9 (4), e1002963. PubMed
Since 2005, I have been working in the realm of molecular biomarker discoveries in Transplantation through my participation in the Genome Canada-funded Biomarkers in Transplantation initiative and with the NCE CECR Prevention of Organ Failure (PROOF) Centre of Excellence. In more recent collaborations with the group, I have developed a computational proteomic pipeline founded on the combination of advanced statistical methodologies to identify, validate, and clinically implement biomarker blood-based solutions to diagnose and predict various diseases. We demonstrated the utility of the proposed methodologies in various biomarkers studies of cardiac transplantation. The computational methodologies proposed in this pipeline provide a reference to a wide range of biomarker proteomic studies. Image from Cohen Freue et al. PLoS Comp Biol 9 (4), e1002963. PubMed
Understanding -omics technologies
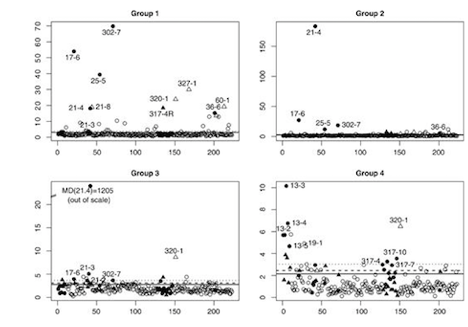 Another important part of my research related with these collaborations has been focused on understanding the design of the experiments, the different sources of variation associated with the technologies used to generate the data, and the quality of the data. My first contribution in this area was the development of MDQC, a novel method based on fundamental concepts of robustness to assess the quality of large-scale microarray datasets. Through more recent collaborations, I have been invited by Dr. Borchers to write an article on the characterization of targeted Multiple Reaction Monitoring (MRM) assays, and to contribute with the design and study of an MRM-MS assay to quantitate putative protein markers of cardiovascular diseases. I am currently leading the development and study of an algorithm, called Protein Group Code Algorithm (PGCA), to link groups of identified proteins (instead of single proteins) across multiple experimental runs. The proposed algorithm addresses an unsolved challenge resulting from the protein inference problem in mass spectrometry data (invited to revise and resubmit in PLoS ONE). Image from Cohen Freue et al. Bioinformatics 23, 3162-3169. PubMed
Another important part of my research related with these collaborations has been focused on understanding the design of the experiments, the different sources of variation associated with the technologies used to generate the data, and the quality of the data. My first contribution in this area was the development of MDQC, a novel method based on fundamental concepts of robustness to assess the quality of large-scale microarray datasets. Through more recent collaborations, I have been invited by Dr. Borchers to write an article on the characterization of targeted Multiple Reaction Monitoring (MRM) assays, and to contribute with the design and study of an MRM-MS assay to quantitate putative protein markers of cardiovascular diseases. I am currently leading the development and study of an algorithm, called Protein Group Code Algorithm (PGCA), to link groups of identified proteins (instead of single proteins) across multiple experimental runs. The proposed algorithm addresses an unsolved challenge resulting from the protein inference problem in mass spectrometry data (invited to revise and resubmit in PLoS ONE). Image from Cohen Freue et al. Bioinformatics 23, 3162-3169. PubMed
Leishmania
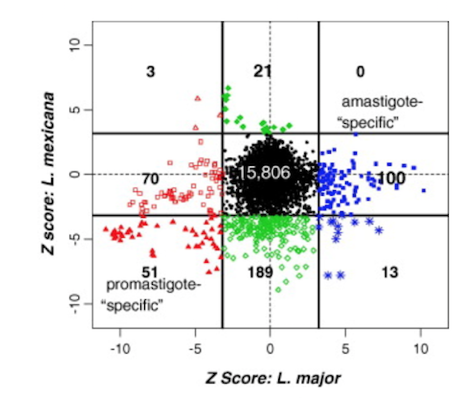 I worked with Dr McMaster, Director of the Vancouver Coastal Health Research Institute, on two studies related to the molecular characterization of different species of Leishmania. Comparative genomic analyses show that the majority of differentially expressed genes between different life stages of Leishmania are species specific. Furthermore, a quantitative proteomic analysis of Leishmania relative protein expression shows a weak correlation to gene expression, suggesting that differential protein modifications may be more important in development than the regulation of gene expression. Image from Cohen Freue et al. Int J Parasitol 37, 1077-1086. link
I worked with Dr McMaster, Director of the Vancouver Coastal Health Research Institute, on two studies related to the molecular characterization of different species of Leishmania. Comparative genomic analyses show that the majority of differentially expressed genes between different life stages of Leishmania are species specific. Furthermore, a quantitative proteomic analysis of Leishmania relative protein expression shows a weak correlation to gene expression, suggesting that differential protein modifications may be more important in development than the regulation of gene expression. Image from Cohen Freue et al. Int J Parasitol 37, 1077-1086. link